Amino acid analysis analysis
|
Protein hydrolysis |
|

|
In order to determine the amino acid content it is necessary to
break the protein chain down into its constituent amino acids by hydrolysis.
The most common conditions for this are treatment with 6N HCl at
110oC for 24 to 72 hours. The optimum time of hydrolysis has to be
determined for each protein as the peptide bonds hydrolyse at different rates.
Most are broadly similar but those between bulky hydrophobic residues take
longer to hydrolyse.
Certain amino acids can be totally destroyed by
hydrolysis if not protected. Tryptophan and tyrosine need to be protected from
chlorine by use of thiol reagents or phenols as scavengers. Cysteine becomes
oxidised on hydrolysis and it is necessary to pre-oxidise to cysteic acid
before hydrolysis. A particular problem is that of deamination of amides -
these are deamidated to the corresponding acidic residues. In this case
determination of the amount of liberated ammonia is needed to quantify the
amines.
|
Identification of amino acids |
|

|
| The amino acids liberated by hydrolysis are
identified and quantified using chromatographic methods. Traditional methods of
analysis involve ion-exchange chromatography and ninhydrin detection, automated
in the amino acid analyser. Current methods can detect down to 50-100 pmol
using ninhydrin and even lower using fluorescamine. |
Modern chromatographic approaches are based
upon HPLC using hydrophobic "reverse phase" columns. Several derivatisation
chemistries are in common use: such as dansyl derivatives,o-pthalaldehyde (OPA)
derivatives, phenylisothiocyanate (PITC) derivatives, or 9-fluorenylmethyl
chloroformate (Fmoc) derivatives. The Fmoc procedure is one of the most widely
used.
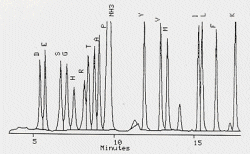 |
| Typical chromatogram of an amino acid analysis |
|
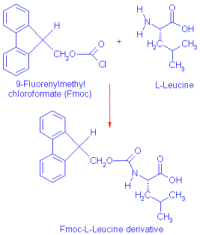 |
| Fmoc derivation of Amino Acids |
|
| Not all amino acids can be
detected with the same sensitivity, different derivatisation chemistries giving
differential sensitivity. For example, OPA-lysine derivatives are unstable and
OPA can not detect proline or hydroxyproline unless they are previously
oxidised with sodium hypochlorite. PITC derivatisation is not very good at
detecting cysteines as cysteic acid and other forms of cysteine resulting from
hydrolysis give poor separation on HPLC of the PTC-derivatives. |
|
Chemical protein sequencing |
|

|
Nowadays it is very common to sequence a protein by the DNA
sequence encoding the protein. This, however, only possible if a cloned gene is
available. It is often the case that chemical protein sequencing must be
carried out in order to generate the oligonucleotide probes needed to clone the
protein. Chemical sequencing can proceed via either the N-terminal or the
C-terminal of the protein. N-terminal sequencing is much more common and is
usually carried out by the Edman procedure.
Fast Atom Bombardment Mass Spectrometry
Fast Atom Bombardment Mass Spectrometry (FAB-MS) is is a very
powerful technique utilising a stream of fast atoms such as argon to achieve
ionisation in the mass spectrometer. This is a "soft" ionisation technique
which generates sequence-specific fragment ions. The sequences of the fragments
can be deduced from their masses. The technique is very sensitive and can
handle mixtures of peptides. The only problem with this technique is its
expense and the equipment is not always available.
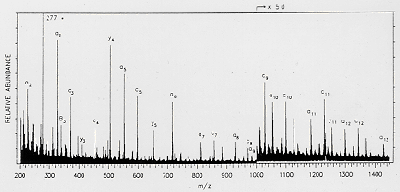 |
| A typical FAB-MS spectrum of a peptide |
Sequencing strategy
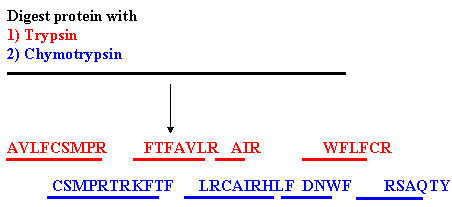 |
| Sequencing strategy using peptides |
|
It is not feasible to sequence an entire
protein molecule. The usual approach is to digest the protein with proteases to
generate peptides and then sequence the peptides. The problem then is to order
the peptides. This is achieved by the use of two peptides which cut the protein
at different points, thus generating overlapping peptides. The peptides are
separated by HPLC and sequenced. The sequence can be deduced from the
overlapping peptides. |
Sequencing from gels
| A very convenient approach to the amino acid
analysis and sequencing of proteins is the use of proteins separated on a PAGE
or IEF gel. In order to perform the analyses, however, the proteins in the gel
must be transfered to a polyvinylidene difluoride (PVDF) membrane. This is
accomplished by electroblotting. |
| The protein bands can then be visualised using
special stains such as Amido Black, Coomasie Blue R-250, Colloidal Gold or
Ponceau S. proteins on the blots can then be hydrolysed by vapour phase HCL
treatment and analysed for their amino acid content by Fmoc derivatives, or
they can be sequenced using automated Edman sequencing directly from the band
on the PVDF membrane. Such sequencing is usually only performed for about 15
cycles or so to generate a "sequence tag". This can then be used to generate an
oligonucleotide probe for cloning or can be used to identify the protein via
sequence analysis. |
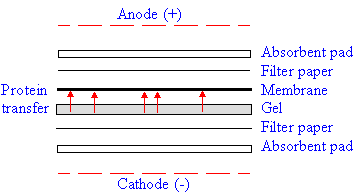 |
| Electro-blotting |
|
|




